User Inputs
Sample files for testing the module can be downloaded from the “Download Sample” link in the red box
- Input 1:
- Protein complex PDB file
- Input 2:
-
Ligand:
- The user may submit the SDF file of the desired ligand in the docked conformation with the protein of interest (docked with the same .pdb file which is uploaded as in the yellow box, Fig. 1).
–OR–
- If the user does not have the docked ligand, they may submit the following three inputs (Fig. 1, purple box):
- Ligand SMILES string
- Protein Chain
- Residue Name (name assigned to the ligand of interest in the uploaded .pdb file)
-
The protein chain and residue names are used to specify the binding site. Hopper uses this information to dock the ligand (given as SMILES) to the protein of interest (given as a .pdb file).
- Input 3:
- Total Number of Molecules to be generated.
- The number of molecules generated will be approximately equal to this number
- Total Number of Molecules to be generated.
- Input 4:
- Job Tag: The user may optionally submit a job tag, which can be used to locate the submitted job in the job queue later
Note: Remember to hit “enter” to submit the tag!
- Job Tag: The user may optionally submit a job tag, which can be used to locate the submitted job in the job queue later
- Submit Job: Once the user has passed these relevant inputs, pressing the “Submit” button submits the job to the queue.
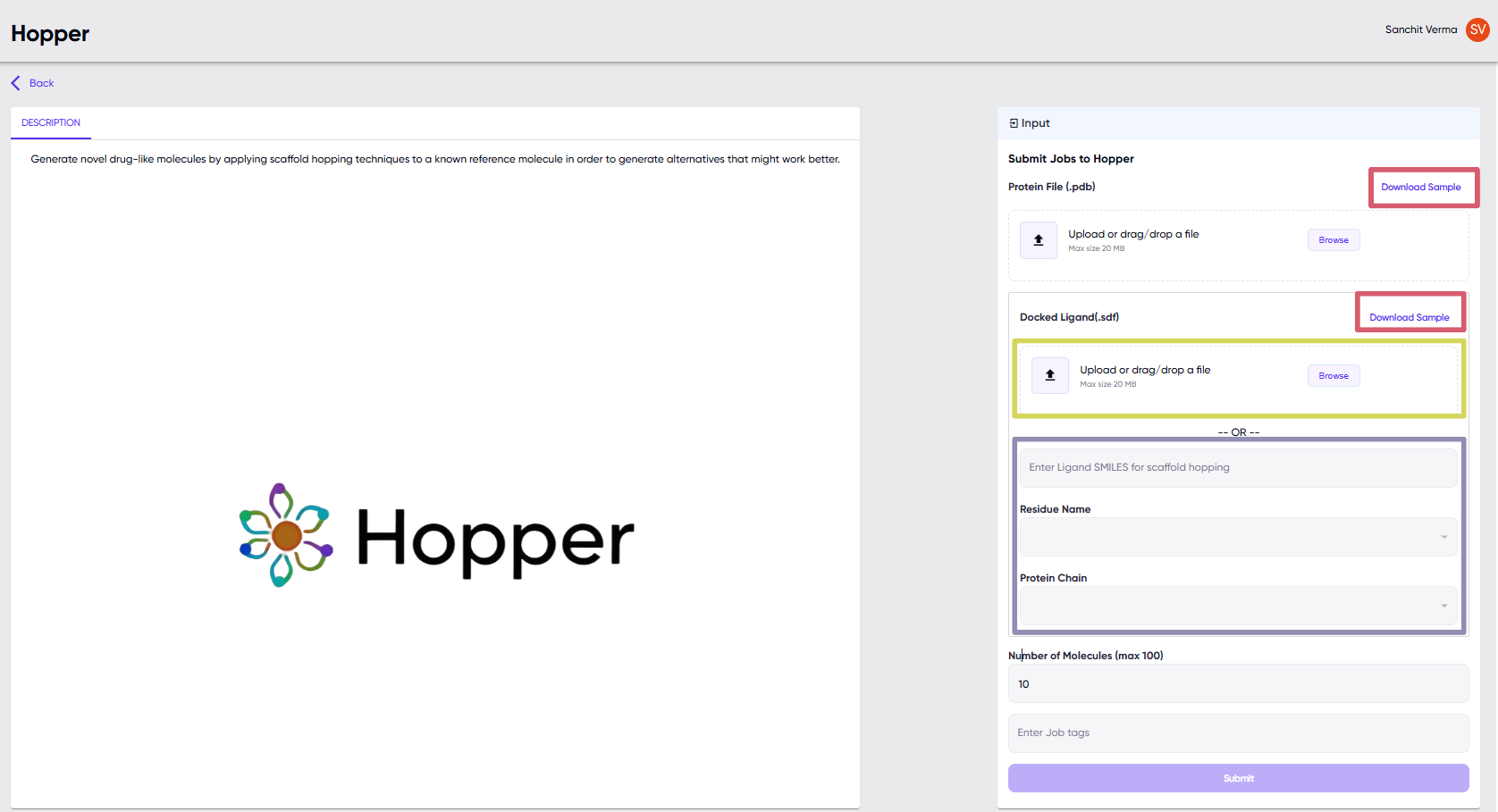
Figure 1: Hopper Dashboard